Class project (Spring 2023)
In this project for my medical image analysis class, I worked with a partner to create a 3D reconstruction of the left ventricle of the heart from ultrasound sequences. This allows for us to estimate the stroke volume and ejection fraction of the heart, which are measures of heart health. The data we were given to work with was a set of 2- and 4-chamber view cardiac ultrasound sequences. These two views are assumed to be orthogonal to each other.
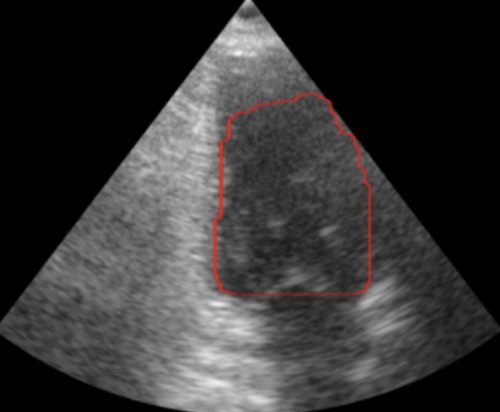
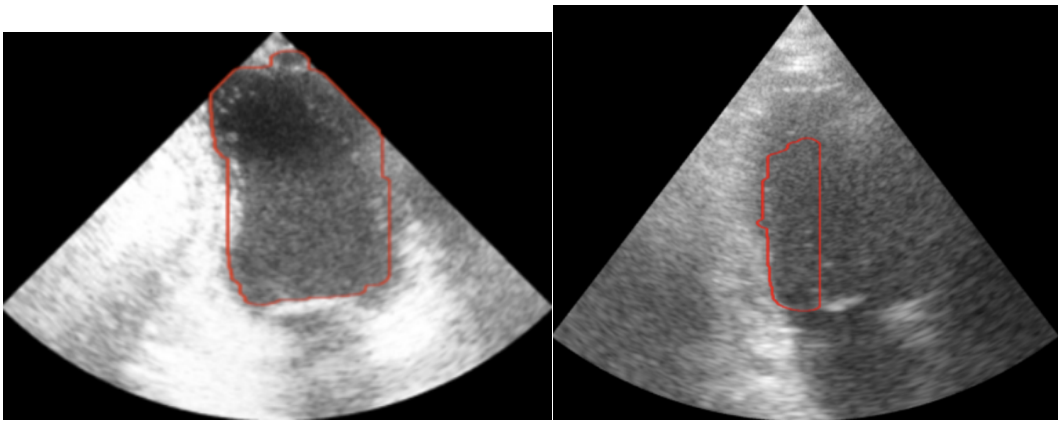
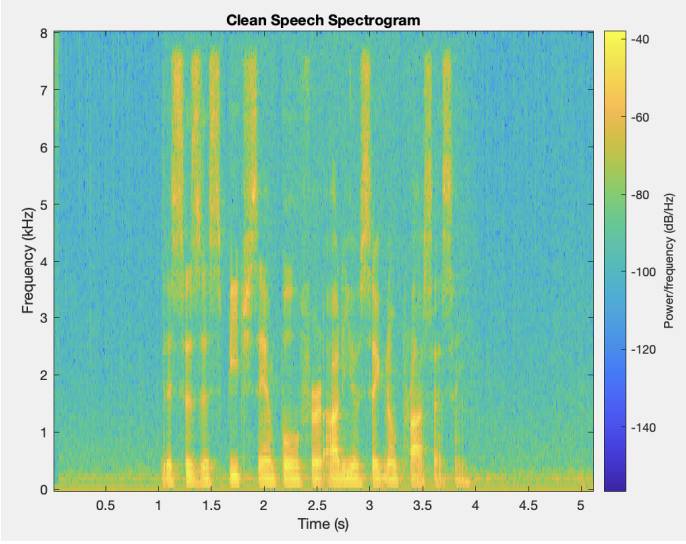
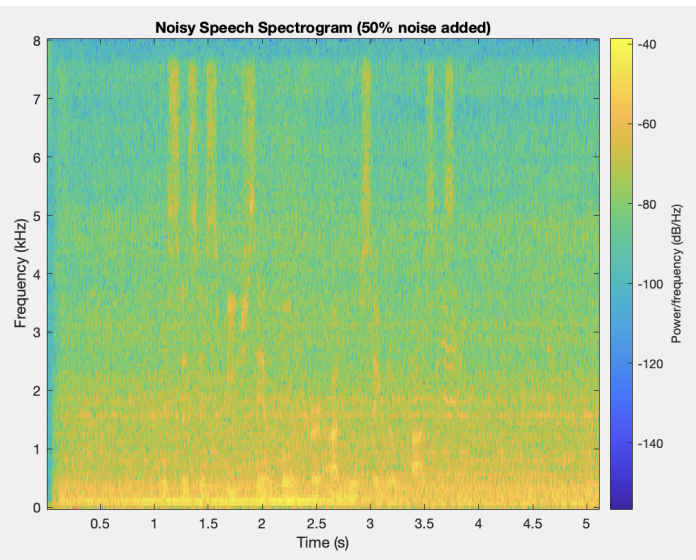
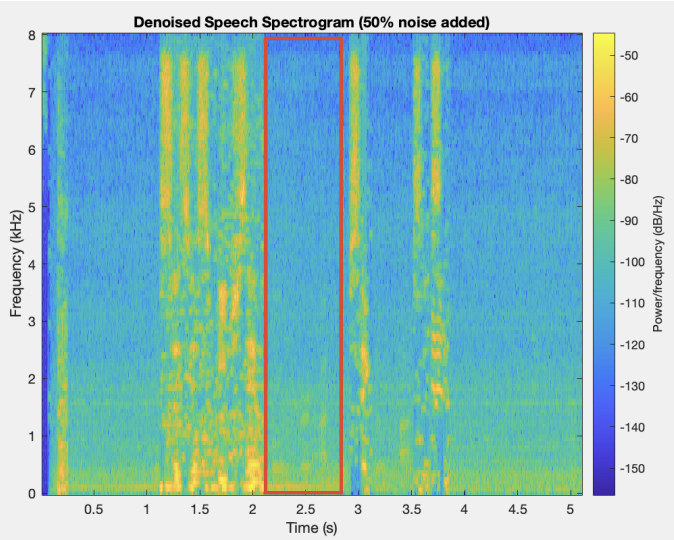
The first step in this project is to segment (find the outline of) the left ventricle. Segmenting the images is the crux of this project because the accuracy of the 3D representation depends entirely on the segmentation accuracy. Other papers used deep learning to segment the ventricle, but we were more interested in using the image analysis techniques we had learned in class. Using a traditional approach to this segmentation problem proved to be extremely difficult because of the poor quality of ultrasound images. We used a variety of techniques to normalize, denoise, and threshold the images, but our results certainly fell short of other groups that used a deep learning approach. Over the entire dataset, our segmentation technique produced a Dice score of 0.6, where a score of 1 corresponds to perfect segmentation.
Due to the nature of this hand-crafted algorithm, the hardest part was obtaining consistent results across all the images in the dataset. As you can see in the example below, some images produce very solid results, whereas others are very poor. Despite this algorithm not performing as well as we had hoped, it was a great challenge for us to apply the techniques we learned in the class.
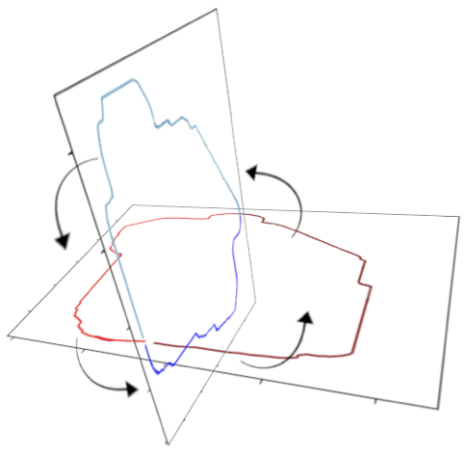
For creating the 3D representation, we leveraged the assumption that the 2- and 4-chamber views are orthogonal to each other. This allows us to essentially interpolate from one set of points to the other set of points to generate points on intermediate angles. See the images below for a visual representation of the algorithm and result.
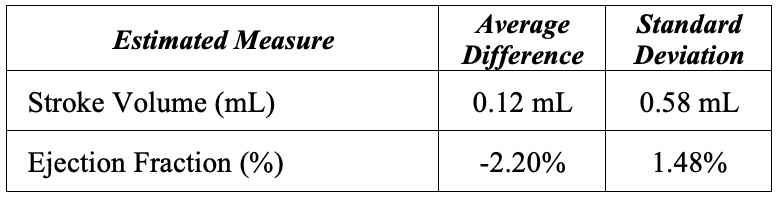
The last part of this project is estimating the stroke volume and ejection fraction for each ultrasound sequence. It was simple to calculate the volume using our 3D surface, but we then had to convert it to mL. Upon plotting our pixel volumes vs physical ground truth volumes, we were able to see that our estimated volumes were not linearly correlated with the physical volumes. Thus, we used an exponential regression to scale our pixel volumes to the physical volumes. See the table below for the average difference and standard deviation over the entire dataset. These results surprised us in how close they were to the ground truth. I would attribute this to the exponential regression correcting the skew produced in our model.
Despite the lackluster results of our algorithm in the segmentation task of this project, I was pleased that we still created an end-to-end algorithm for a useful measure of heart health that’s based off of an uninvasive ultrasound. This project challenged me to be creative, not only in the image processing methods but also creating a 3D model from two 2D images.